In melanoma tissue, the proportion of positive cells for genes related to cell death (PRGs) GZMA, GSDMB, NLRP1, IL18, and CHMP4A is lower than in normal skin. Cellular pyroptosis is a new field that affects the tumor microenvironment and tumor immunotherapy. However, the role of cell pyroptosis is still controversial, partly due to the heterogeneity of cell composition in melanoma.
In August 2023, Professor Li Bin's team from the Institute of Dermatology at Shanghai Academy of Traditional Chinese Medicine published a research paper titled Clinical mediated discovery of pyroptosis in CD8+T cells and NK cells reveals melanoma heterogeneity by single cell and bulk sequence in the journal Cell Death&Disease.
This article provides a comprehensive analysis of the single-cell transcriptome of melanoma specimens. We found that the expression of PRGs is dysregulated in immune cells, such as CD8+cells (representing CD8+T cells) and CD57+cells (representing NK cells). In addition, immunohistochemistry and multiple immunofluorescence staining experiments further confirmed that GZMA+cells and GSDMB+cells are mainly expressed in immune cells, especially in CD8+T cells and NK cells.
In melanoma specimens, the presence of GZMA+combined with CD8+T cells (0.11%) and GSDMB+combined with CD57+cells (0.08%) was minimal, compared to 4.02% and 0.62% in the control group, respectively. These findings suggest that a decrease in immune cells in tumors may reduce the ability of cells to undergo pyroptosis, thereby posing a potential risk in combating the characteristics of melanoma.
We constructed a prognostic risk model and personalized prediction model based on single-cell and whole RNA seq analysis (C-index=0.58, P = 0.002),PRGs may play a role in the prevention of malignant melanoma. In summary, the identification of immune cell populations and immune gene modules through experimental verification helps us better understand cell pyroptosis in melanoma.
In this article, the researchers used the TissueFAXS Spectra panoramic multispectral tissue scanning quantitative analysis system from TissueGenomics to obtain images. Obtain images and perform quantitative analysis using StrataQuest software.
Panel 1 :DAPI,CD8,GZMA,GSDMB
Panel 2:DAPI,CD57,GZMA,GSDMB
Although the impact of cell pyroptosis on cancer is still well established, there are still extensive unknowns waiting to be explored in research on cell pyroptosis.Considering the close relationship between the study of cell pyroptosis and the inflammatory response process, with the help of TissueFAXS Cytometry technology and multi-color immunofluorescence staining, not only can cells with specific protein expression related to pyroptosis be accurately identified, but also high-throughput, high-precision, and informative quantitative analysis of the spatial distribution, morphological characteristics, and interactions with other cell types of pyroptosis levels in tissues can be achieved.
At the single cell quantitative level上,firstly, cells are counted by identifying nuclear markers, and then the morphology of cytoplasm/membrane staining is accurately identified using the nuclear expansion algorithm of Tissue Cytometry. After obtaining the contour area of single-cell true staining, statistical analysis is performed on the pixel intensity of each cell protein marker to ultimately obtain the true intensity level of single-cell protein expression. This method is used for precise screening and division of positive thresholds, and even further identifies the functional relationship between positive cells and adjacent negative cells.
In this article, the author also utilized the approach of multi omics research to propose new insights into the cell-cell interaction network related to the pathogenesis of melanoma. Using CD8 positive T cells as research clues, the imbalance of CD8 cell edge infiltration was observed, further expanding the breadth of research in the field of tumor immunity mechanisms. In addition, in situ precise spatial quantification analysis was conducted on NK cells, GZMA cells, and GSDMB cells, laying a solid foundation for further in-depth research. In these studies, multiple immunohistochemical markers in situ were used to analyze protein expression and cell distribution of cells with different phenotypes, and individualized precise quantitative analysis was performed using TissueFAXS Cytometry technology.
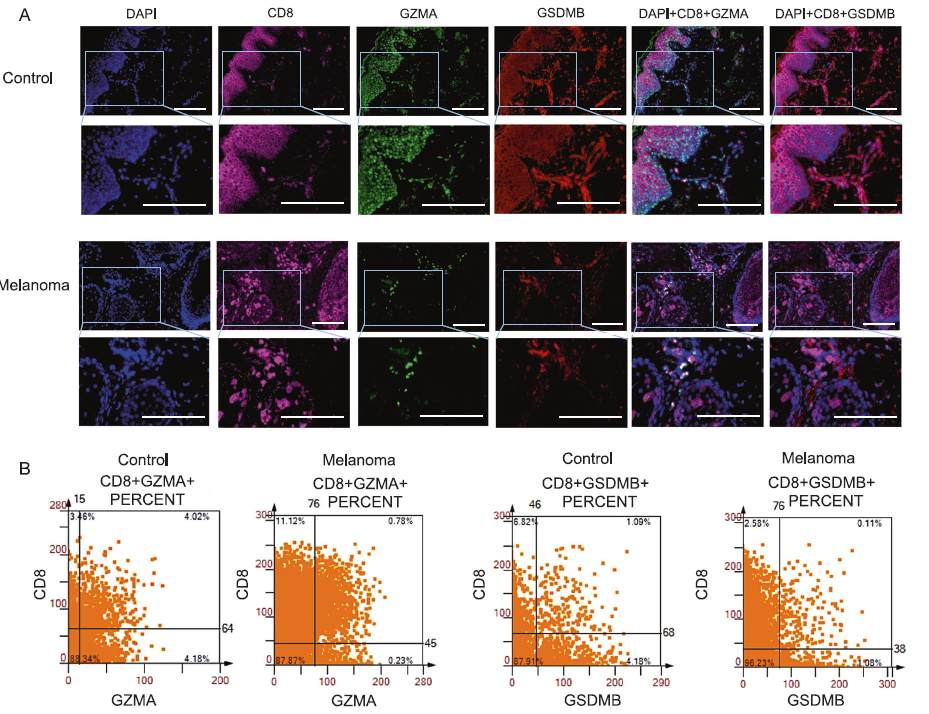
Figure 1 GZMA+cells and GSDMB+cells are secreted by CD8+T cells.
A:Multi color immunofluorescence staining images of control group and melanoma tissue. DAPI (blue), CD8 (pink), GZMA (green), and GSDMB (red).
B:Scatter plots of CD8+GZMA+co localization and CD8+GSDMB+co localization.
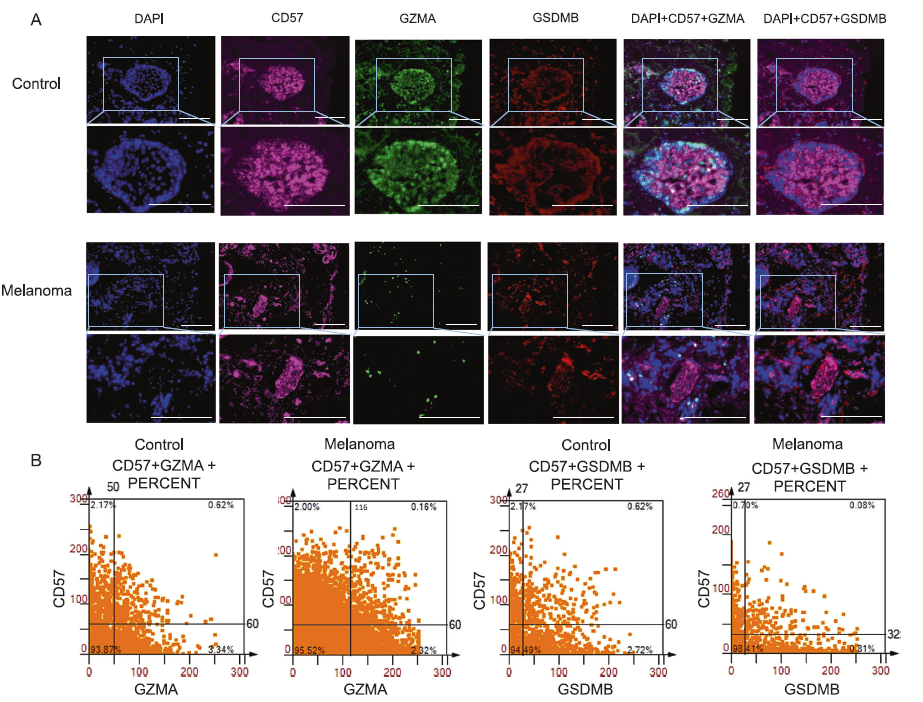
Figure 2 NK T cells secrete GZMA+cells and GSDMB+cells
A:Multi color immunofluorescence staining images of control group and melanoma tissue. DAPI (blue), CD57 (pink), GZMA (green), and GSDMB (red).
B:CD8+GZMA+co localization, CD8+GSDMB+co localization, CD57+GZMA+and CD57+GSDMB+scatter plots.