Lung cancer is the leading cause of cancer-related deaths worldwide, resulting in approximately 1.8 million deaths annually, with lung adenocarcinoma (LUAD) being the predominant pathological type. The tumor microbiome is an important component of the tumor microenvironment, and its interaction with the host is crucial for the occurrence and development of cancer. Therefore, research on tumor microbiota provides a new perspective for the diagnosis, screening, and treatment of cancer. Although the 'multimodal microbiome' has become one of the hallmarks of cancer, little is known about the role of the fungal community within tumors as living microorganisms in cancer progression.

On September 21, 2023, Wang Hui/Liu Ningning from the School of Medicine of Shanghai Jiao Tong University, Zhang Peng from Tongji University, and Chen Changbin's research group from the Pasteur Institute of the Chinese Academy of Sciences jointly published an article titled "The intratumoral mycobiome promoters lung cancer progress via myeloid derived suppressor cells" in the top international oncology journal Cancer Cell (IF=50.3).
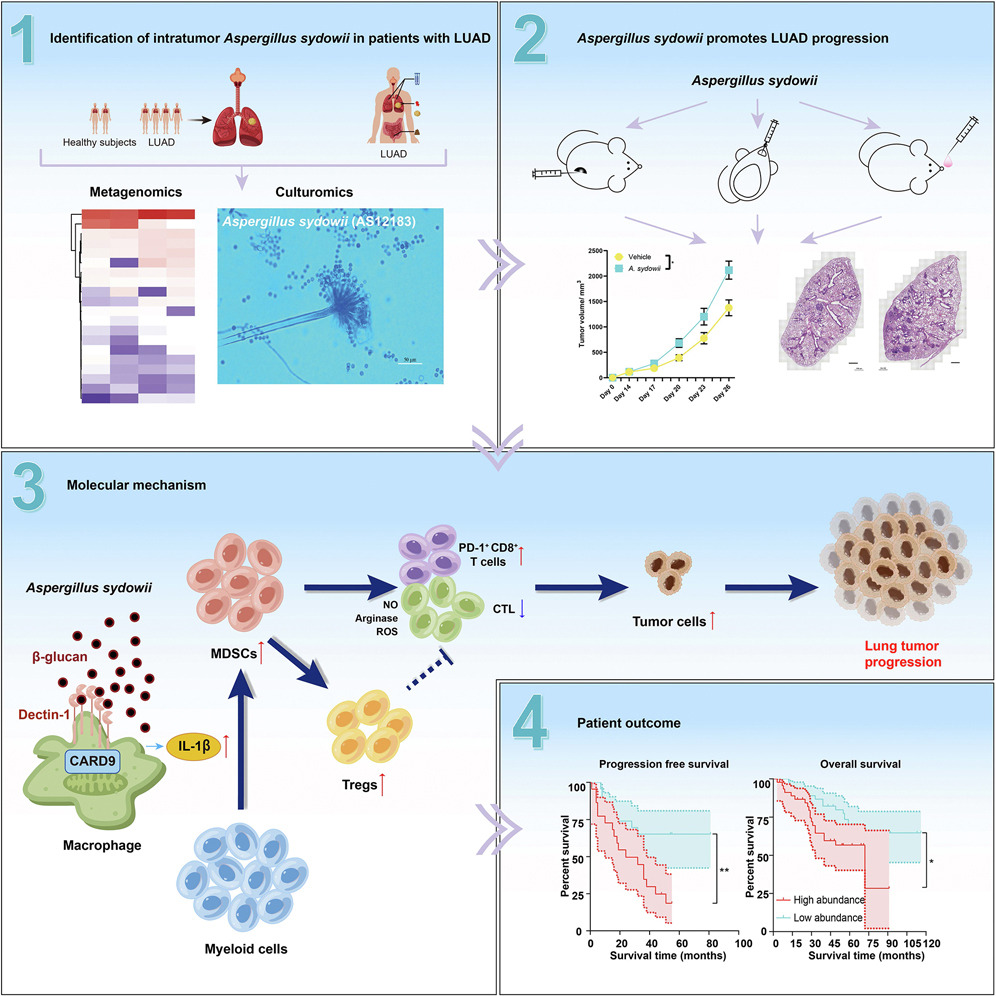
This article used DNA extraction rich in fungi and deep metagenomic sequencing to discover the enrichment of tumor resident Aspergillus nidulans A. in lung adenocarcinoma (LUAD) patients sydowii。 Through three different homologous lung cancer mouse models, we found that Pseudomonas aeruginosa inhibits the activity of cytotoxic T lymphocytes and promotes the accumulation of PD-1+CD8+T cells through IL-1b-mediated expansion and activation of bone marrow suppressor cells (MDSCs), thereby forming an immunosuppressive tumor microenvironment that promotes lung tumor progression. This is achieved through the secretion of IL-1b mediated by the b-glucan/lectin-1/CARD9 pathway. The analysis of human samples confirms the enrichment of A Sydowii is associated with immune suppression and poor patient prognosis. The research results indicate that the fungal community within tumors, despite having low biomass, can promote the progression of lung cancer and serve as a potential target for improving the prognosis of LUAD patients.
Experimental part
Compared to general research on cancer, the author of this article focuses on the impact and mechanism of fungal dysbiosis on cancer, and uses FISH staining technology to label the target microbial community. According to the research needs, the urgent issues to be addressed are twofold: firstly, to understand the relationship between microbiota and tumors from a macro perspective; secondly, to analyze the mechanism of action between fungi and cells from a micro perspective. In order to conduct in-depth exploration of the above two issues, the author used StrataQuest quantitative analysis software to establish precise quantitative analysis standards.
Due to technical principles, FISH staining imaging results often exhibit high background and low signal states. StrataQuest quantitative analysis software, as a powerful tool for data analysis in Tissue Cytometry technology, also has its unique analysis strategy for spot fluorescent signals labeled with FISH staining: firstly, in terms of recognition accuracy, it can accurately identify each signal point under high background and count the molecules of nucleic acid fragments; Secondly, based on the identification and counting, combined with the localization of the cell nucleus and the cytoplasmic contour localization of a certain protein staining, the spatial localization of nucleic acid expression was achieved at the microscopic level, and data on the role of fungi and their related cells could be obtained simultaneously. This method can also be used for consistency localization analysis of certain specific gene replication, transcription, and translation processes, which is extremely valuable for verifying the transitivity of central dogma.
At the macro level, Tissue Cytometry technology can also identify tumors and freely define the spatial location of the microenvironment around the tumor to analyze the spatial distribution relationship of single cells with different phenotypes within the tumor microenvironment. Through simple data statistics, the interaction relationship data that previously required bioinformatics analysis algorithms can be obtained, such as the interaction between microbiota and tumors, or the interaction between different inflammatory cells in the tumor microenvironment.
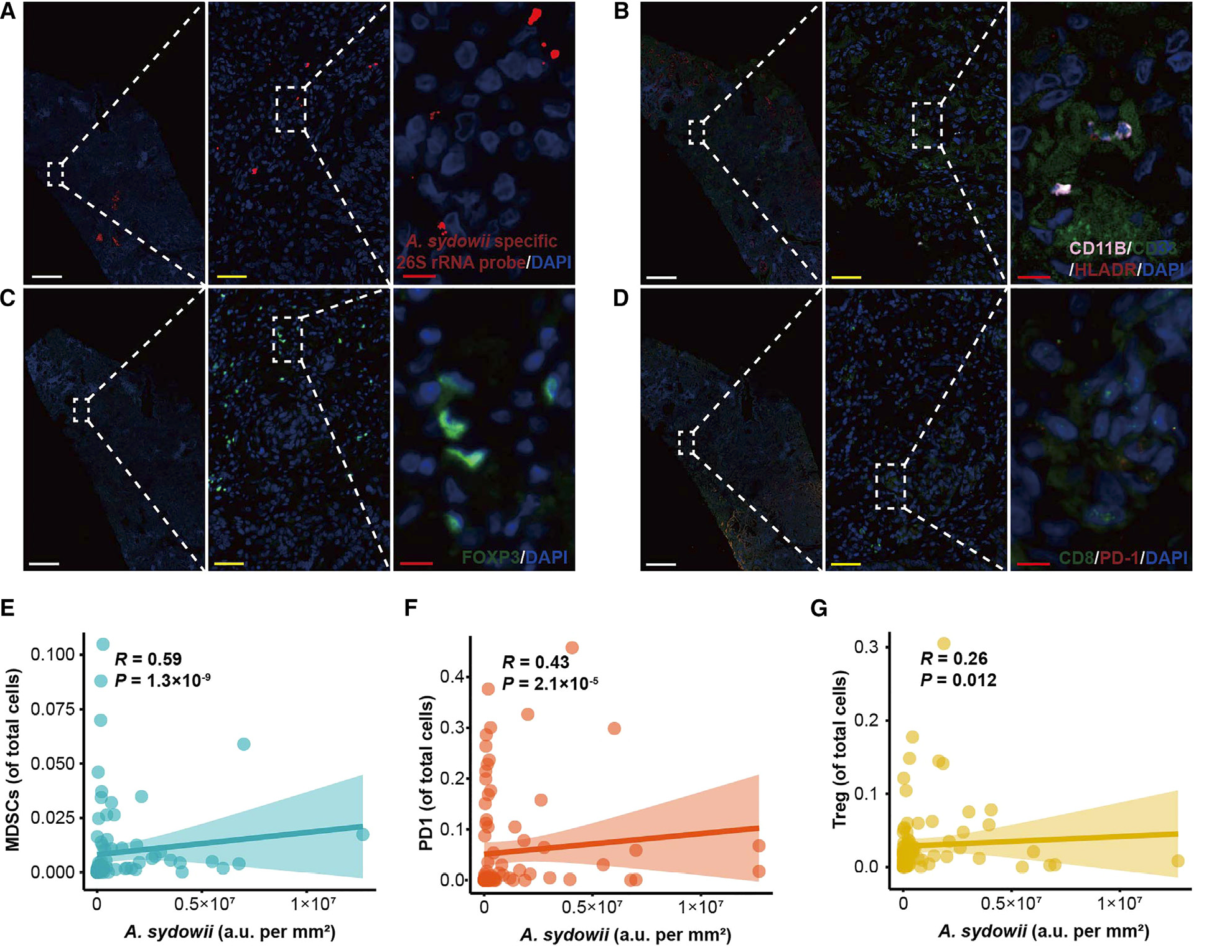
Figure 1 A.The high abundance of sydowii is associated with immune suppression and poor patient prognosis
(A)Multi color immunofluorescence of clinical samples from LUAD patients shows A. sydowii and A. sydowii specific FISH images within the tumor. red A.Sydowii; Blue, DAPI。
(B)Multi color immunofluorescence images of MDSCs within tumors from clinical samples of LUAD patients (CD11b+CD33+HLADR -). Pink, CD11b; Green, CD33; red HLADR; Blue, DAPI。
(C)Multi color immunofluorescence images of tumor associated Tregs (FOXP3+) in clinical samples of LUAD patients. Green, FOXP3; Blue, DAPI。
(D)Multi color immunofluorescence images of PD-1+CD8+T cells (PD-1+CD8+) within tumors in clinical specimens of LUAD patients. Green, CD8; red PD-1; Blue, DAPI。
(E-G)The correlation between the abundance of A. sydowii in tumor tissue samples of LUAD patients and MDSCs (E), Tregs (F), and PD-1+CD8+T cells (G).
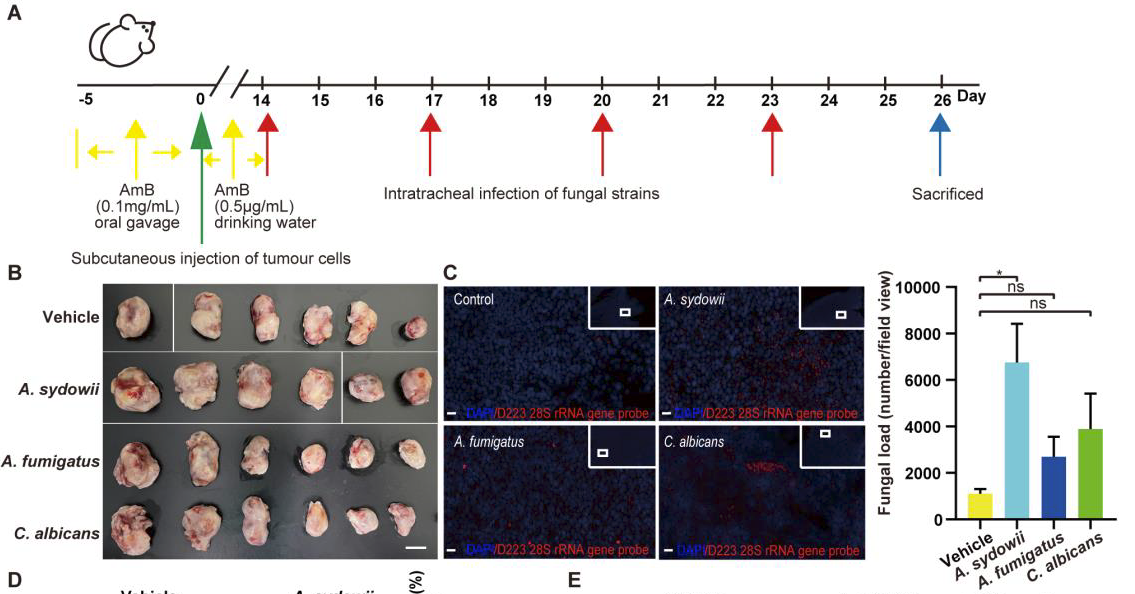
Figure 2 A.Sydowii was unable to promote the immunosuppressive activity of M2 macrophages.
(C)Comparing the abundance of fungi in tumors of mice infected with A.111 sydowii, Aspergillus fumigatus, Candida albicans, or vector by FISH staining with D223 28S rRNA probe in subcutaneous LLC mouse models. Red fungal FISH probe; Blue, DAPI。