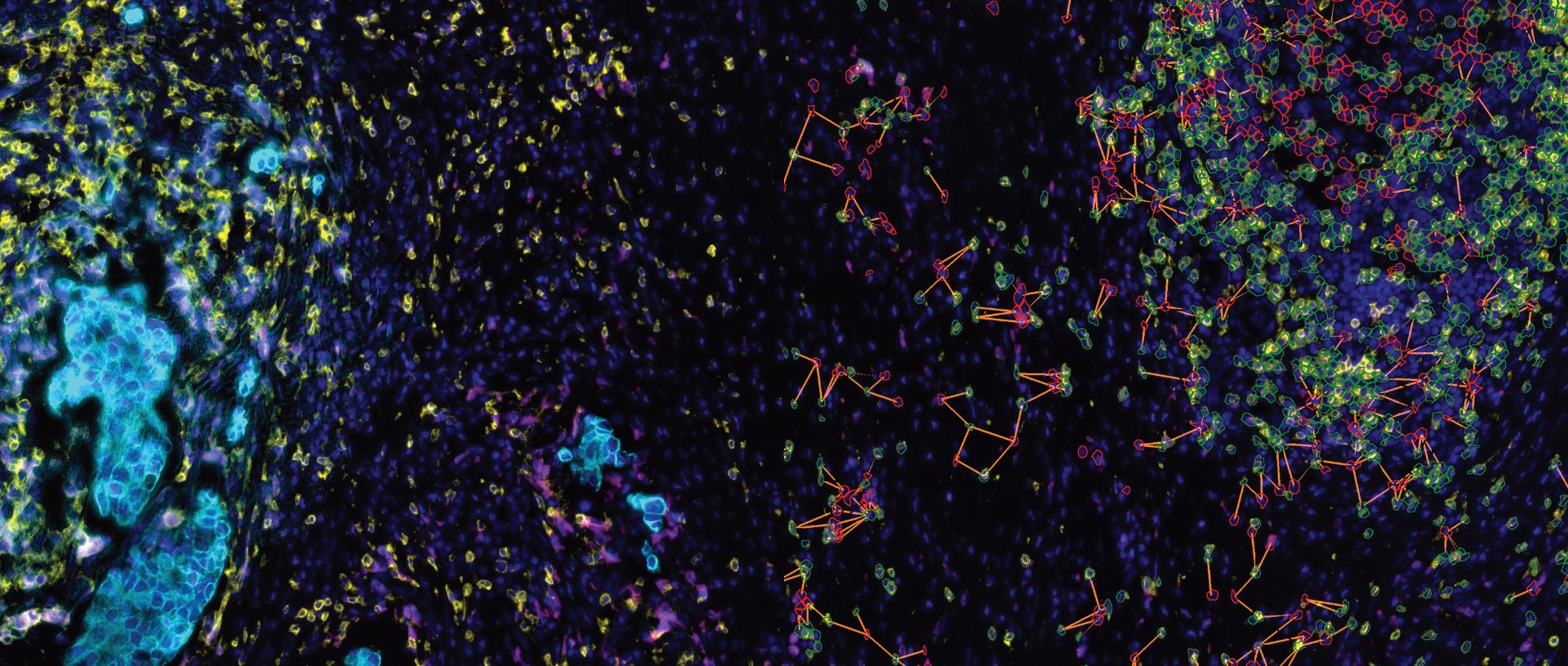
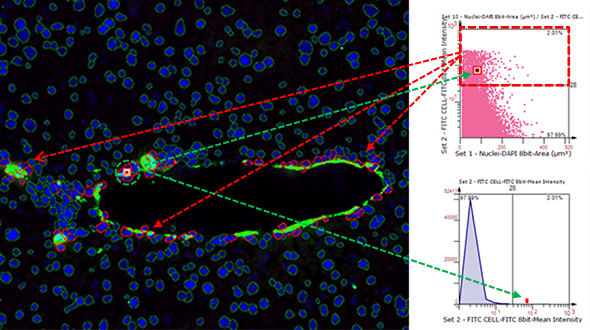
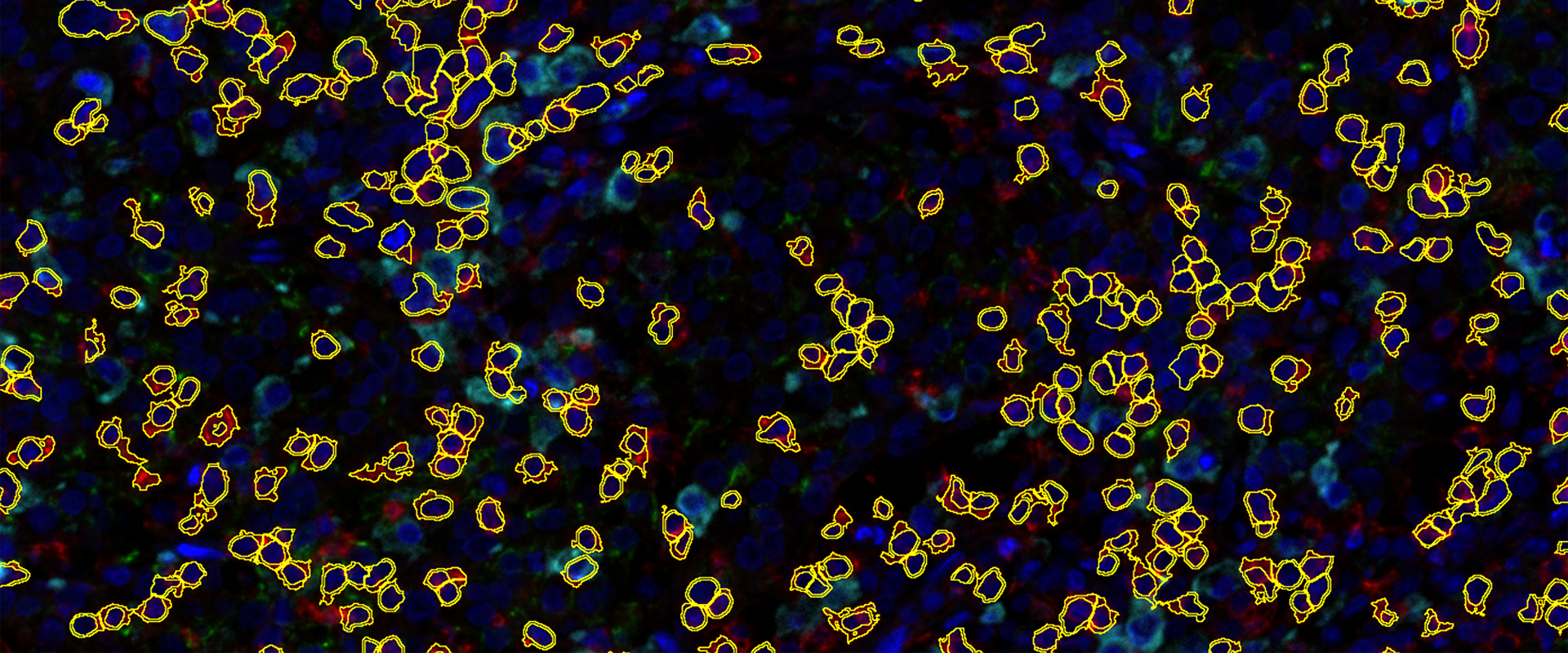
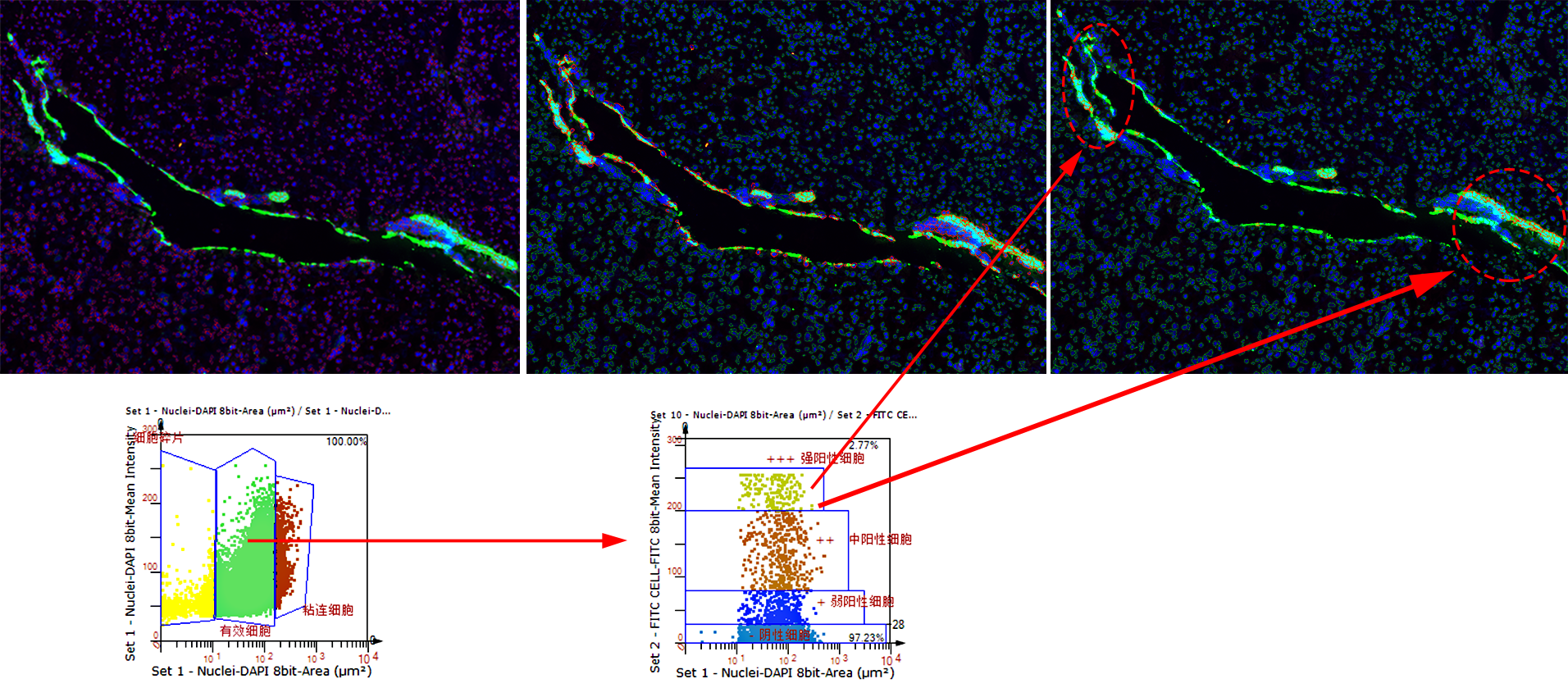
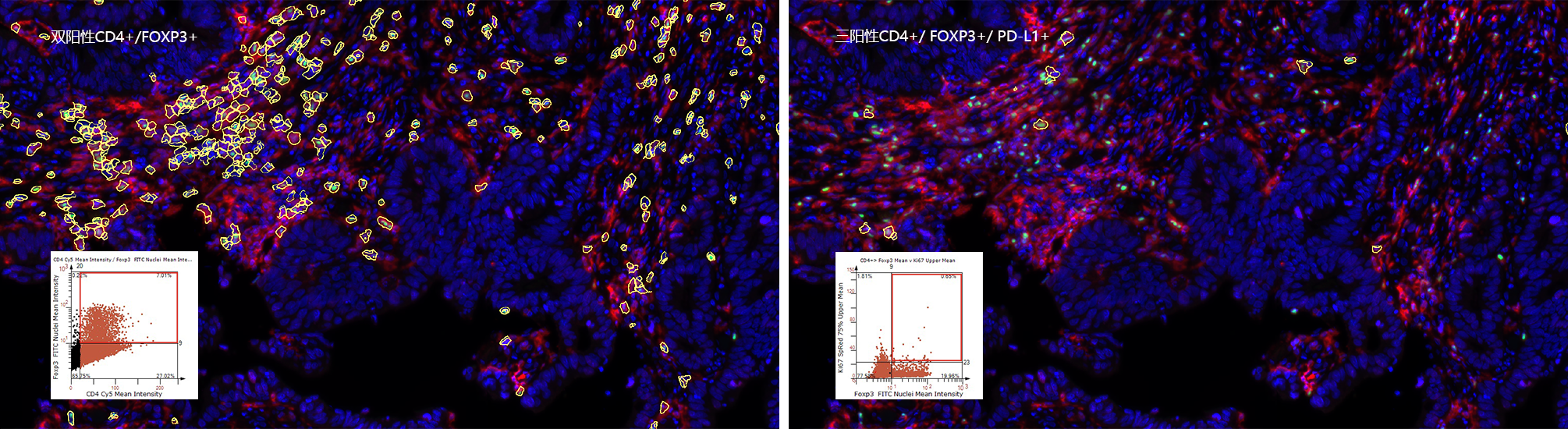
TissueQuest (TQ) is a software platform for the automatic identification and quantification of single cells in situ in tissues, which is used to solve the screening of specific staining signals on nucleus/plasmid/membrane of different types of cells in fluorescent samples. Based on the staining condition of the samples and the morphological characteristics of the cells, it can accurately analyse the differences in data between different experimental groups.
TQ adopts the streamlined operation mode of sample import + one-click analysis, and then adjusts the analysis parameters based on the results of quantitative data analysis by using the scatterplot forward and backward retrospective tool to adjust the analysis parameters according to the differences of the samples. This iterative data acquisition process, based on the powerful image computing capability of the software, obtains the big data information contained in the slices in real time, and guides the researcher to finally obtain the data results that reflect the actual situation of the samples.
Nucleus recognition algorithm
Multiple cytoplasmic morphology recognition modes
In situ analysis of cellular phenotypes
Forward and reverse backtracking
FISH analysis
Multiple data presentation modes