Severe combined immunodeficiency (SCID) is a genetic disease that can lead to severe deterioration of the immune system. Among the key genes related to SCID, RAG1 and IL2RG play a crucial role. IL2RG is essential for the development, differentiation, and function of T, B, and NK cells, while RAG1 plays a crucial role in adaptive immunity by promoting V (D) J recombination during lymphocyte maturation. Animal models carrying these gene mutations exhibit significant immune system deficiencies.
Non human primates (NHPs) are ideal models for biomedical research due to their genetic and physiological similarities with humans. Base editing technology (CBE) is a powerful tool for accurately and effectively modifying single base mutations in the genome. Their successful implementation has been demonstrated in human cells, mice, and crop species.

On September 4, 2023, the team led by Yan Sen and Tu Zhuchi from the Hong Kong Macau Institute of Central Nervous Regeneration at Jinan University published a research paper titled "Generation of Inactivated IL2RG and RAG1 Monkeys with Severe Combined Immunodeficiency Using Base Editing" in Signal Transmission and Targeted Therapy (IF=39).
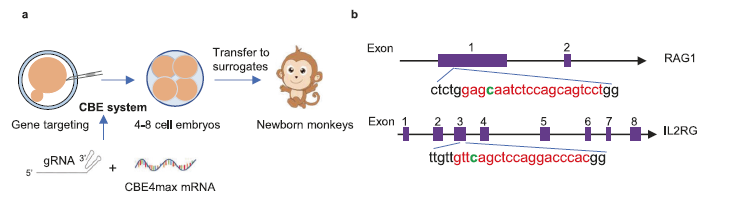
This article provides an overview of the use of the CBE4max system to establish an immunodeficient monkey model by inactivating the IL2RG and RAG1 genes. Monkeys that have undergone base editing exhibit severe immune system damage, characterized by decreased lymphocytes, atrophy of lymphoid organs, and lack of mature T cells. In addition, these base editing monkeys can carry and support the growth of human breast cancer cells, thus leading to the formation of tumors.
In summary, we have successfully developed an immune deficient monkey model with the ability to promote tumor growth using the CBE4max system. These immunocompromised monkeys demonstrate enormous potential as valuable tools for advancing biomedical and translational medicine.
Experimental part
This article uses the TissueFAXS plus panoramic tissue cell quantitative analysis system from TissueGenomics to collect images and analyze in situ single-cell data of thymus and spleen tissue slices from base edited monkeys.
In order to evaluate the feasibility of using base edited monkeys as an in vivo model for studying human oncology, researchers used Tissue Cytometry technology to accurately quantify key phenotype cells in the thymus and spleen of wild/mutant monkeys, and utilized its precise quantitative characteristics to confirm with WB and flow cytometry, further improving the reliability of the data.
Compared with traditional immunohistochemistry and immunofluorescence techniques that evaluate protein expression levels based on staining intensity, the Tissue Cytometry technique first identifies the cell nucleus and filters out adherent cells and cell debris through tissue flow cytometry. Then, using in situ real single cells as the core, the complete contour of the target protein staining is delineated. Finally, by calculating the intensity and contour of the complete staining of single cells, the precise quantitative target of single cells is achieved.
Recently, more and more scholars have combined multi omics analysis techniques and finally chosen Tissue Cytometry as a validation method for single-cell data. With the help of the spatial interaction analysis method of Tissue Cytometry, they have further expanded the analysis of tissue in situ sections not only as the ultimate validation method, but also as a solution for in situ spatial omics analysis.
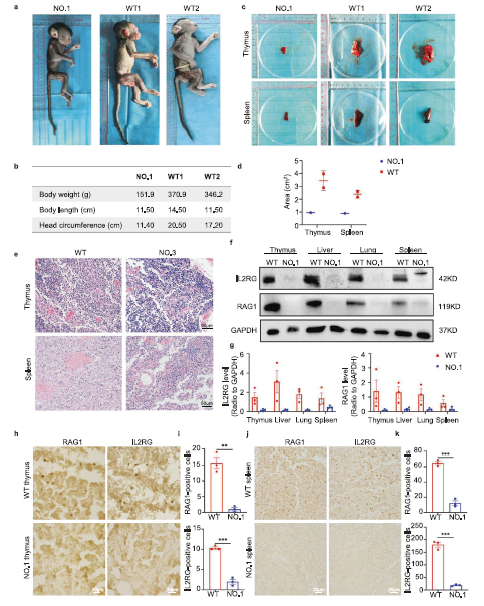
Figure 1 Phenotypic analysis and histopathological changes of base edited monkeys
e:HE images of thymus and spleen development in base edited monkeys
h-j: Immunohistochemical imaging and quantitative analysis of RAG1 and IL2RG in thymus and spleen of mutant monkeys and WT monkeys
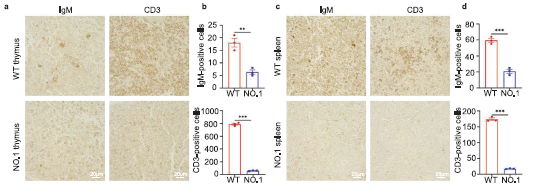
Figure 2 Research on T and B lymphocytes of base edited monkeys
a-d:Immunohistochemical images and quantitative analysis of IgM+and CD3+in thymus (a, b) and spleen (c, d)
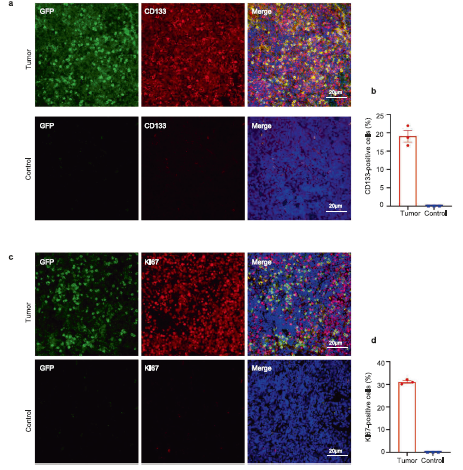
Figure 3 In situ validation of tumor tissue
Ki67 and CD133 labeled tumor tissue immunofluorescence images and quantitative analysis